Homosexual behaviour in humans is, historically, a controversial topic. People who exhibit these behaviours are commonly subject to discrimination and abuse; to this day, rights for LGBT+ people is still hotly debated around the world. In the 1990s, a scientific paper emerged claiming that researchers had discovered a genetic marker for homosexuality – the so-called ‘gay gene’. This whipped up a storm of media attention and ignited the debate about nature vs. nurture when it comes to same-sex behaviour. Attempts to replicate this study have usually failed, implying it could be an outlier.
After 22 years, the concept of a ‘gay gene’ may have finally been put to rest by a paper published in 2019. Researchers have attempted to characterise the genetic makeup of same-sex behaviour using a study involving 477,522 people – the most powerful study in its field to date.
The research involved what is known as a ‘Genome-wide association study’ (GWAS). This is an observational study where scientists compare DNA to certain traits, telling them if the traits are caused by specific genes. Among other findings, the study concluded that same-sex behaviour is not caused by a single gene, but instead caused by many small variations across multiple genes that have an additive effect to a specific trait. Scientists describe this as being ‘polygenic’ behaviour.
The study used genetic data from sources like 23andMe and the UK Biobank. Studies on this scale this have only been made possible due to the advent of widespread genetic screening companies. The genetic data was cross referenced with questionnaires about the subject’s sexual history.
The study focused on single nucleotide polymorphisms (SNPs). These are variations in the bases on DNA found between humans.

The researchers found that there are 5 significant genome-wide signals responsible for same-sex behaviour. These are the SNPs present in a high enough proportion of people that show this behaviour. Despite finding five significant signals, these only account for roughly 8-25% of same-sex behaviour, implying there are many more genes at play here contributing in tiny ways to a person’s sexual behaviour.
Another interesting aspect is that, out of the five signals, two of them occurred exclusively in males and one of them occurred only in females. Researchers suggest that the partially different genetic factors in men and women could indicate different hormonal influences on sexual behaviour. For example, the importance of testosterone and oestrogen in sexual attraction.
As a follow up to these results, researchers explored genetic correlations between same-sex sexual behaviour genetics and 28 pre-selected traits. This was to identify if any overlap existed between them. Several traits were identified, among them were cannabis usage, risk-taking behaviour, openness to experience and major depressive disorders.
Finally, the researchers looked exclusively at the people who reported same-sex behaviour and investigated whether the proportion of same-sex partners had any correlation with the genetic variable. They found that there was no correlation between the proportion of same-sex partners and the same-sex behaviour variable. This suggests that the genetic variable for same-sex behaviour and the proportion of same-sex partners are separate on a genetic level.
This completely reframes how society views sexuality. Instead of it being a binary scale from ‘heterosexual’ to ‘homosexual’, it may be more accurate to view heterosexuality and homosexuality as separate traits that people have. Traditionally, the ‘Kinsey Scale’ has been used to place people’s sexual preferences. The scale implies that the more attraction people feel towards the same sex, the less they will feel to the opposite sex – which the study refutes.
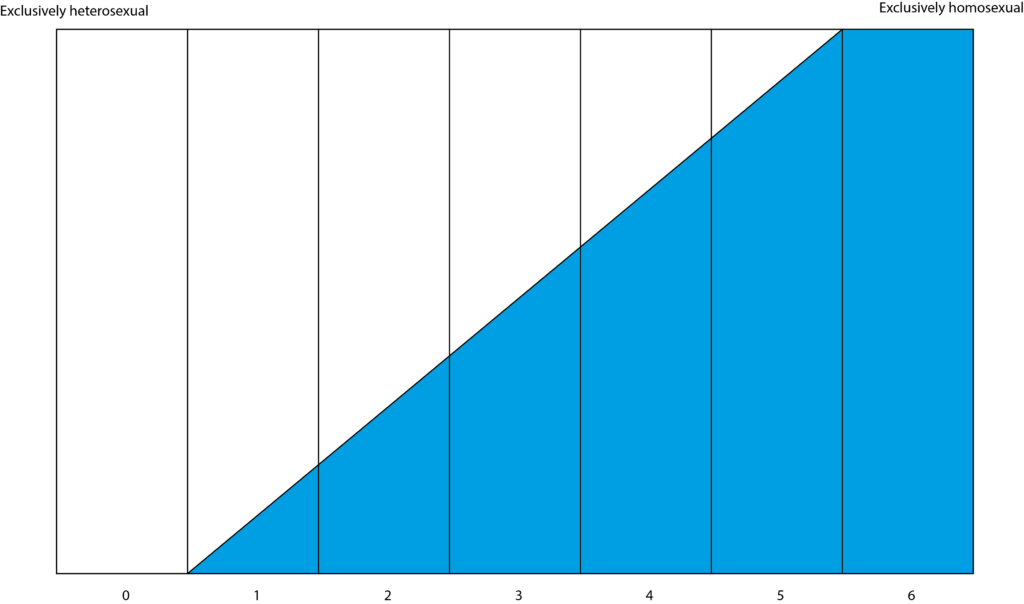
Adopting a different scale that is more reflective of human genetic architecture will also allow bisexuality and asexuality to be expressed more clearly. Using separate measures of same-sex and opposite-sex attraction could go a long way to helping people understand themselves more.
